Multi-scale and longitudinal data modelling in pharmacology: toward digital pharmacological twins
Coordinating Partners : Jean-Baptiste Woillard and Christophe Battail
Coordinating institution : Inserm
Pharmacology, multi-scale, digital pharmacological twins, therapeutic drug monitoring, pharmacogenomics, molecular modelling, dose individualization, transplantation, anti-infectious, oncology
Translational pharmacology is a discipline seeking to move results from molecular pharmacological research to the clinic in order to optimize the choice of drugs, their dosage and safety in patients. Specifically, it seeks to translate knowledge acquired from molecular and preclinical models to the bedside patient studies.
Two pillars in pharmacology are:
- the drug target, involving the interaction between the drug and its target and the effect of the drug at a microscopic scale (e.g. molecular inhibition) and at the macroscopic scale (e.g. positive or dverse effect),
- the blood/plasma drug concentration, involving many factors of variability such as genomics pharmaco-genomics and pharmaco epigenomics), inflammation, drug-drug or drug-food interactions (including harmacokinetic and pharmacodynamic interactions), organ dysfunction, and individual (e.g. circadian rhythms and age).
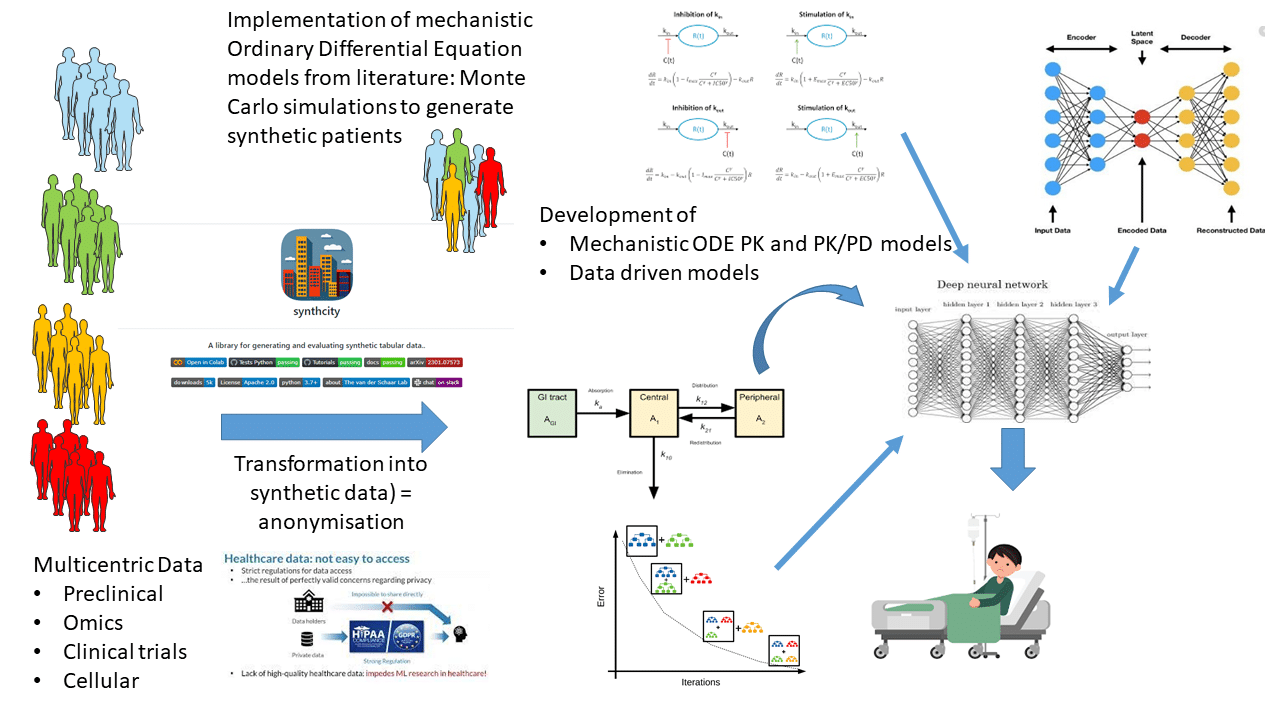
All these aspects are associated with methodological challenges that have to be overcome based on mechanistic and statistical modelling (including machine learning approaches). This also requires the integration of longitudinal bservation and time-dependent confounders. During the last decade, large amounts of preclinical and patient data has been generated, including omics data.
Some of these have been mathematically modelled using mechanistic approaches. However, the community of pharmacologists and methodologists does not overlap either at the level of the model scale (molecular, tissue, patient), or at the level of the model type (drug interactions, pharmacokinetics, pharmacogenomics…), preventing a global approach to the personalization of drug therapies. Structuring this multi-disciplinary community under the notion of systems pharmacology requires investment in the development of interoperable models at each scale of space and time.
The project includes 4 work packages:
- organisation (WP1),
- collection of multiscale and multisource data and models (WP2),
- inference of models at each biological scale up to the patient level (WP3)
- building meta-models (WP4).
These meta-models will act as “digital pharmacological twins” and will be used:
- before treatment, to guide treatment selection (drug combination, dose and concentration target) or to explain special dose requirement, based on individual features at multiple scales and
- during treatment, by integrating dynamic data in order to refine the therapeutic strategy.
Three case studies corresponding to current challenges in pharmacology will be used to develop the digital pharmacological twin framework:
- transplantation,
- infectious diseases
- oncology.
| Laboratory or department, Teams | Supervisors |
| P&T – U 1248
BGE – UA13 |
Inserm, Limoges University, CHU Limoges
Inserm, Grenoble Alpes University, CEA |
| Idesp – UA11, Team PreMeDICaL | Inserm, Inria, Montpellier University |
| Inserm UMRS 1138 – Inria Paris Team HEKA | Inserm, Inria, Paris Cité University |
| IRSET – UMRS 1085 Rennes, Team DRIVE | Inserm, Rennes University, EHESP |
| Oncopole Claudius -Regaud- ICR/Oncopole
CRCT – U 1037 Inserm/ UMR 5071 CNRS – Team Dose Individualization of Anticancer Drugs -DIAD |
Inserm, CNRS, Toulouse 3 University
Institut Universitaire du Cancer de Toulouse – Oncopole partner |
| CRCM – U1068 / UMR7258 –
Team COMPO |
Inserm, Inria, CNRS, AMU
Institut Paoli-Calmettes partner |
| CRT2I – U1064 et UR1155 | Inserm, Nantes University,
CHU Nantes partner |
| Laboratoire HP2 – U1042 | Inserm, Grenoble-Alpes University,
CHU Grenoble partner |